Student Wiki on methodology
PLEASE: DO NOT change the INDEX page !!!
This page contains the links to the nine official subjects, which are the same in the Choice.
To contribute, go to the correct page by clicking on the description here in the index, then click EDIT and contribute. At the end, please save.
IMPORTANT !!!
Please do not make extensive cut-and-paste: it s useless, anybody can go to the source you use and read it. Read the texts, digest, and make a short résumé. If you wih you can include link(s) to the source(s).
Other contributors can revise, add, erase, modify... Please do not repeat the same text as well.
RNA binding protein mapping: RIP, CLIP, HITS-CLIP, PAR-CLIP
(Restore this version)
Modified: 27 March 2020, 11:30 AM User: Stefano Romanazzi →
Back to index
[Stefano Romanazzi]
RNA IMMUNOPRECIPITATION (RIP)
This technique is very similar to ChIP: a specific antibody is used to pull down the RNA binding proteins(RBP) together with their associated RNA, it can be used both for mRNA and non-coding RNA; isolated RNA can then be futher analyzed with other techniques, like RT-PCR or sequencing.
There are two main RIP variants:
Native RIP: direct identification of the bound RNA and the protein of interest in the immunoprecipitated sample, this approach typically allows recovery of high affinity protein-RNA interactions
Cross-linked RIP: live cells are treated with formaldehyde first, to generate RNA-proteins cross-links, this allows precise mapping of the direct and indirect binding site of the protein to the RNA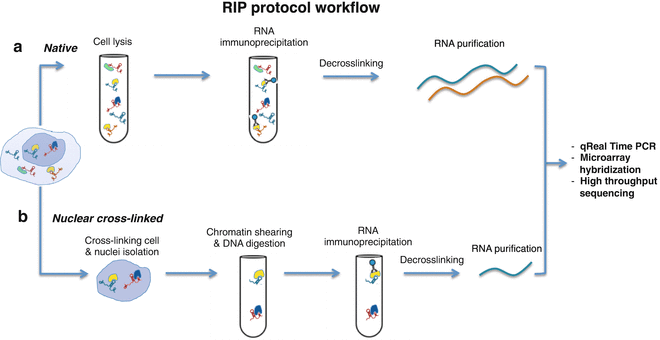
PROTOCOL SUMMARY
1. Cell harvesting (and optional treatment with formaldehyde)
2. Nuclei isolation
3. Chromatin shearing
4. RNA immunoprecipitation
5. Washing off unbound material
6. Purification of RNA that was bound to immunoprecipitated RBP
