RNA binding protein mapping: RIP, CLIP, HITS-CLIP, PAR-CLIP
(Restore this version)
Modified: 19 March 2018, 7:39 PM User: Lucia Giorgi →
RNA binding protein mapping
RNA-protein interaction is important in controlling many aspects of gene regulation. In fact, different proteins are required for mRNA processing, such as splicing. What’s more, the properly cellular functions of several non-coding RNAs (ncRNAs) depend upon the formation of RNA-protein complexes.
To identify these interactions, different techniques have been developed. The predominant ones are ‘protein centric’, which require the knowledge of the protein which is analysed. They are based on the purification of proteins followed by the sequencing of RNA molecules associated with them, in order to map the RNA binding proteins (RBPs) across the transcriptome.
RNA immunoprecipitation (RIP)
A first example is the RNA immunoprecipitation (RIP) technique. This method is an antibody-based technique which purifies RNA-protein complexes under physiological conditions. The protein of interest is immunoprecipitated together with its associated RNA, which can then be purified, reverse transcribed in cDNA, amplified with PCR and sequenced.
The advantage of this technique is that the native complexes present in the cells are preserved. Yet, this kind of technique has some limitations, between them the most concerning is the possibility of formation of non-physiological RNA-protein interactions in solution, in fact the re-association of molecules after cell lysis is possible.
Cross-linking immunoprecipitation (CLIP)
To distinguish between in vivo interactions
and interactions that form in solution, RNA-protein complexes are cross-linked
in cells, in order to create covalent linkage between physically interacting RNA and proteins. In the CLIP technique this is done using short
wavelenght UV light. Then, RNA-protein complexes are purified using stringent wash conditions followed
by denaturation of all complexes by heating in sodium dodecyl sulphate
(SDS), running the samples on an SDS-polyacrylamide gel electrophoresis
(PAGE) gel, and extracting the crosslinked RNA-protein complex, which
will run at a size slightly larger than the protein itself, from the gel.
(Alessia Fucini)
HITS-CLIP
HITS-CLIP is an evolution of CLIP technique, it is a crosslinking immunoprecipitation (CLIP) coupled with high throughput sequencing (HITS-CLIP). It is used to understanding about how protein-RNA complexes interactions regulate gene expression in living cells.
The original CLIP experiments showed that CLIP was able to identify direct protein-RNA interactions, and that such interactions identified functionally relevant points of RNA-protein interaction. However, the small number of tags precluded drawing any robust generalizations about the nature of the RNA-protein interactions. This limitation was overcome by applying next-generation high throughput sequencing methods to CLIP, termed HITS-CLIP. Analysis of the same RNA-protein interactions with HITS-CLIP yielded over 1000-fold more unique tags for the same cost.
In the figure is reported the HITS-CLIP procedure: (a) UV irradiation of live cells or tissue induces RNA-protein cross-links (Steps 1–3). (b) Material is lysed, RNA is partially digested and the target protein is immunopurified with antibody coupled to magnetic beads (Steps 4–17). (c) Alkaline phosphatase treatment removes 3′ hydroxyl to permit 3′ linker ligation (Steps 18–20). (d) Radiolabeled (*) 3′ linker (red) is ligated to RNA tags (Steps 21 and 22). (e) Polynucleotide kinase treatment phosphorylates 5′ RNA ends, allowing subsequent 5′ linker ligation (Steps 23 and 24). (f) Complexes are eluted from beads and separated by SDS-PAGE. After transfer to nitrocellulose membrane, complexes are visualized by autoradiography (Steps 25–32). (g) RNA is extracted from the desired membrane region by protease treatment, and the 5′ linker (purple) is ligated to tags (Steps 33–52). (h) Tags are amplified by RT-PCR (Steps 53–77). (i) After the addition of sequencing adapters in a second PCR step, the samples are sequenced on the Illumina platform (Steps 78–87).
(Elena Doria)
PAR-CLIP
To improve the cross-linking efficiency, photoactive nucleoside analogues, 4-thio-uridine (4-SU) and 6-thioguanosine (6-SG), and hence photoactivatable ribonucleoside-enhanced CLIP (PAR- CLIP) were used by Tuschl and colleagues. In PAR- CLIP, 4-SU and 6-SG are added to the growth medium, which are then taken up by cells and eventually incorporated into newly-synthesized RNA molecules without obvious toxicity. The formation of covalent crosslinks between proteins and RNAs is performed under UV irradiation at 365 nm, instead of UV 254 nm used in CLIP. The following steps are similar to CLIP protocol, including RNase treatment, immunoprecipitation, recovery of RNA fragment, reverse transcription and sequencing. It is noteworthy that the incorporated 4-SU can lead to T to C transition in the sequenced cDNA. Therefore, it is possible to identify crosslink sites at individual-nucleotide resolution by analyzing the mutations in cDNA sequences
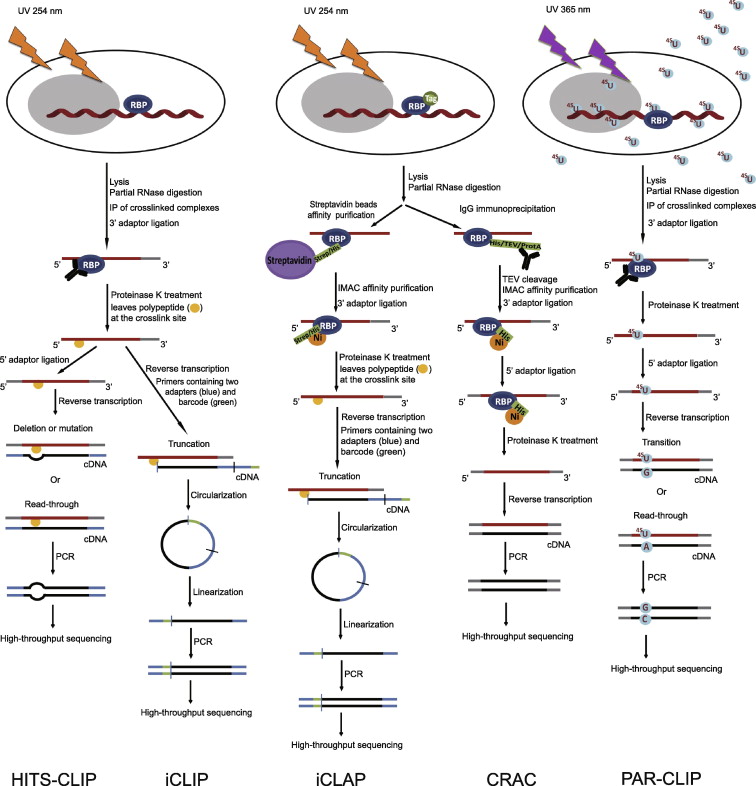
FIgure 1: Outline of HITS-CLIP, PAR-CLIP and several variants, iCLIP, iCLAP and CRAC.
High-throughput sequencing CLIP (HITS-CLIP) and individual-nucleotide resolution CLIP (iCLIP) are in the left panels; individual- nucleotide resolution crosslinking affinity purification (iCLAP) and crosslinking and cDNA analysis (CRAC) are in the middle panels; photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) is in the right panel. PAR-CLIP uses thioribonucleosides and UV at 365 nm to form the complex of RNA and RNA-binding protein (RBP), while the other four methods utilize UV at 254 nm. Isolation of RNA–RBP complexes is achieved either by immunoprecipitation (IP) (PAR-CLIP, HITS-CLIP and iCLIP) or by double affinity purification (iCLAP and CRAC). iCLAP and CRAC use immobilized metal ion affinity chromatography (IMAC) under denaturing conditions as a secondary purification. To achieve individual-nucleotide resolution, HITS-CLIP utilizes deletion or mutation during reverse transcription, iCLIP and iCLAP take advantage of truncated cDNAs, and PAR-CLIP makes use of thymidine (T) to cytidine (C) transition in cDNA. TEV, tobacco etch virus; ProtA, Staphylococcus aureus protein A.
Reference 1: http://www.sciencedirect.com/science/article/pii/S1672022914000230
(Lucia Giorgi)
