Long-range chromatin interactions: 3C and derivatives up to Hi-C
(Restore this version)
Modified: 18 March 2018, 7:56 PM User: Riccardo Aucello →
3C
Chromosome conformation capture (3C) technology and its derivatives have been widely used to analyze the spatial organization of chromatin in a cell. These methods quantify the number of interactions between genomic loci that are nearby in 3-D space, but may be separated by many nucleotides in the linear genome. It can be applied to investigate the nuclear juxtaposition of any two genomic regions, in cis or trans. Such interactions may result from biological functions, such as promoter-enhancer interactions, or from random polymer looping, where undirected physical motion of chromatin causes loci to collide. 3C provides information on 3D chromatin structures that occur in living cells.
The protocol involves formaldehyde cross-linking of chromatin fragments and proteins in proximity followed by chromatin isolation and digestion with a restriction enzyme. The specific enzymes used are chosen in order to free a known or predicted DNA-DNA interaction mediated by a protein complex. The freed fragments are then ligated into rings and the crosslinks are reversed and the DNA is purified. Proteins are removed through the exposition to high ionic strands or high temperature. Both end-point PCR and quantitative real-time PCR (qPCR) can be employed to quantify the abundance of purified DNA 3C ligation products. The abundance of these recombinant fragments directly correlates to the interaction frequency of the two ligated regions.
This basic principle can be combined with other technologies to increase scale or specificity of the DNA loops being interrogated.
The derivative techniques of 3C are:
- Circularized Chromosome Conformation Capture (4C)
- Carbon Copy Chromosome Confromation Capture (5C)
- Chromatin interaction analysis by paired-end tag sequencing (CHIA-PET)
- ChIP-loop
- Hi-C
4C
As told before, 4C means Circularized Chromosome Conformation Capture. The first part of the protocol is equal to the one for basic 3C, so it will be reported without modification from the previous entry (tanks to the author).
The protocol involves formaldehyde cross-linking of chromatin fragments and proteins in proximity followed by chromatin isolation and digestion with a restriction enzyme. The specific enzymes used are chosen in order to free a known or predicted DNA-DNA interaction mediated by a protein complex.The freed fragments are then ligated into rings and the crosslinks are reversedand the DNA is purified. Proteins are removed through the exposition to high ionic strands or high temperature.
Following, these ligated segments are treated with another RE (a 4 pb RE), to create sticky ends: so, the two extremities will be able to interact with each other providing circularization. PCR is then carried out with divergent primers, preferentially recognising the contact points of the two fragments; sequencing can be managed after this passage.
(Samuele Irudal)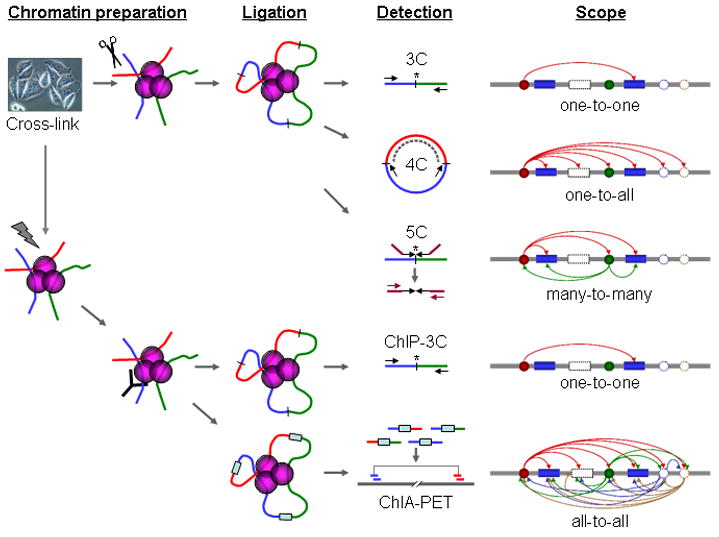
Fig 1. Schematic comparison of 3C, 4C, 5C, ChIP-3C, and ChIA-PET
ChIP-3C has been developed for a more specific identification of chromatin interactions, it detects DNA-DNA interactions mediated by a specific protein of interest. ChIP-3C is the result of combination between ChIP to the 3C protocol with the goal to reduce the non-specific noise that characterizes both these methods. The main advantage of ChIP-Loop is it reduces the background noise in 3C experiments and increases the specificity by selecting for a known protein mediating the DNA-DNA interaction. A reduction in background noise over a simple 3C assay is achieved by removing a large amount of genomic DNA by IP. Further, by targeting analysis to a specific protein of interest only specific, biologically relevant interactions are detected.
ChIP-3C has yielded insights into chromatin looping as mechanisms whereby important proteins can mediate cellular functions such as gene regulation. ChIP and 3C were first combined in 2005 to study a mouse model of Rhett. These researchers found that formation of a silent-chromatin loop is important for the function of the Dlx50Dlx6 locus. Further, they found that this loop was mediated by MECP2 and that this interaction is lost in individuals with Rhett Syndrome.
There are a few different ChIP-3C protocols in use, one of these involves:
· preparation of urea ultracentrifugation-purified
· restriction enzyme-digested
· cross-linked chromatin
· ChIP enrichment
· proximity ligation
· reverse cross-linking to free DNA fragments from protein binding
· detection using PCR
.
While another omitted the urea ultracentrifugation purification and simply combined 3C and ChIP:
· cells are are formaldehyde cross-linked followed by chromatin isolation and restriction digestion
· Cross-linked chromatin is immunoprecipitated using an antibody against a protein of interest
· the sticky DNA fragment ends are ligated while the chromatin fragments are still bound to the antibody,
· The cross-links are reversed and the DNA is purified. A specific DNA-DNA interaction of interest can then be detected using PCR
However, the same final result is that predicted DNA-DNA interactions that are mediated by a protein of interest are confirmed.
Another variant on ChIP-3C is the so-called 6C technique, which uses a cloning approach for detection, instead of site-specific PCR employed in conventional ChIP-3C.
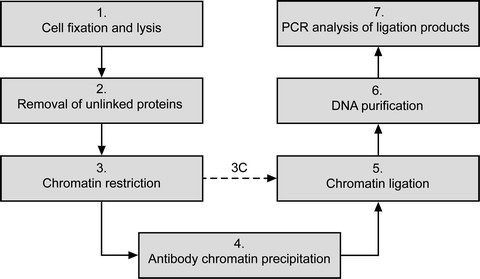
Fig 2. The main steps of ChIP-loop assay
One complication of is the accurate quantification of interaction levels, which must take into account both ChIP enrichment of the sites as well as high levels of non-specific chromatin noise due to random. Because 3C contains much noise, and does not include any steps to separate specific interactions from non-specific interactions before detection; therefore, if anyone uses ChIP to pull down specific protein-bound chromatin interactions from such 3C chromatin fragments, one would also co-precipitate the non-specific chromatin fragments that are attached to the specific interaction complexes. As such, it is likely that the use of the standard 3C protocol with the addition of ChIP just as it is would lead to high levels of false positives.
Minimize ligation bias by having sufficient starting material. By performing the ligation when the DNA is concentrated in a small volume, undesirable cross-ligation can occur between chromatin fragments. Validate with ChIP alone if possible. Cross-referencing ChIP-loop with ChIP data greatly reduces false-positives in the ChIP-loop assay. Use appropriate controls:
- Make a control templete that contains ligation products in equal amouts.
- Determine interaction frequency between known loci of increasing distance to estimate random interaction.
- When comparing two conditions, determine interaction frequency for a region expected to be the same in the two states.
Debate on the efficacy ChIP-3C rises questions on what is the nature of non-specific chromatin interactions, how such non-specific interactions are different from true and specific interactions, and how to experimentally separate non-specific chromatin interactions from true ones. The practical question is: can the non-specific and specific interactions be physically separated? If such a method can be found, the reduction in noise would greatly benefit the field of chromatin interactions.
(Riccardo Aucello)