NGS-genome sequencing
Table of contents
1. 1. Illumina sequencing [edit]
1.1. Introduction to NGS
1.1.1.
The Basics of NGS Chemistry
2. 2. Paired-end sequencing [edit]
3. 3. Ion torrent sequencing [edit]
5. 5. Pacific Bio and Nanopore long-run sequencing [edit]
5.1.1. PacBio long-read sequencing
5.1.2. Nanopore sequencing
Return to index
Tentative paragraphs:
(Dallorto Eleonora)
1. Illumina sequencing [edit]
Introduction to NGS
Next generation sequencing (NGS), massively parallel or deep sequencing are related terms that describe a DNA sequencing technology . NGS is considered the catch-all term used to describe a number of different modern sequencing technologies. Using NGS an entire human genome can be sequenced within a single day. In contrast, the previous Sanger sequencing
technology, used to decipher the human genome, required over a decade to deliver the final draft. The short read, massively parallel sequencing technique has been a fundamentally different approach, which revolutionized sequencing launching the “next generation” in genomic science.
Illumina® sequencing technology was developed by Shankar Balasubramanian and David Klenerman of Cambridge University, later founder of Solexa, a company acquired by Illumina®. (Go for a brief history of Illumina® sequencing https://emea.illumina.com/science/technology/next-generation-sequencing/illumina-sequencing-history.html?langsel=/it/)
Illumina® sequencing is based on the incorporation of reversible dye terminators that enable the identification of single bases as they are incorporated into DNA strands.
This technique offers some advantages over traditional sequencing methods such as Sanger sequencing. The automated nature of Illumina® sequencing makes it possible to sequence multiple strands at once and obtain actual sequencing data quickly. The innovative and flexible sequencing system enables a broad array of applications in genomics, transcriptomics, and epigenomics.
The Basics of NGS Chemistry
A DNA polymerase catalyses the incorporation of fluorescently labelled deoxyribonucleotide triphosphates (dNTPs) into a DNA template strand during sequential cycles of DNA synthesis. During each cycle, at the point of incorporation, the nucleotides are identified by fluorophore excitation. The critical difference is that, instead of sequencing a single DNA fragment, NGS extends this process across millions of fragments in a massively parallel manner.
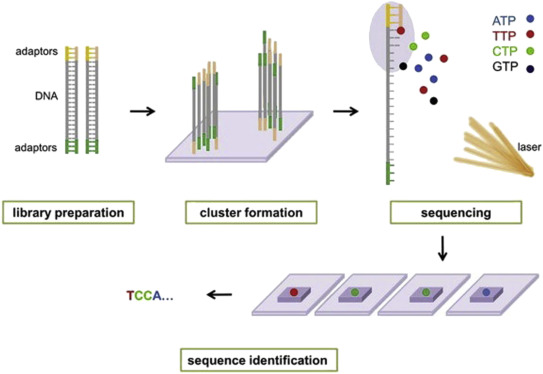
Illumina® sequencing technology works in four basic steps:
- Library Preparation: The first step is to break up the DNA into more manageable fragments of around 200 to 600 base pairs. Short sequences of DNA called adaptors are attached to the DNA fragments. Alternatively, “tagmentation” combines the fragmentation and ligation reactions into a single step: enzymes called transposases randomly cut the DNA into short segments ("tags"), which greatly increases the efficiency of the library preparation process. Adapter-ligated fragments are then PCR amplified and gel purified.
- Cluster Generation:The library is loaded into a flow cell where fragments are captured on a lawn of surface-bound oligos complementary to the library adapters. The complementary DNA binds to primers on the surface of the flowcell and DNA that does not attach is washed away. Unlabeled nucleotide bases and DNA polymerase are then added to lengthen and join the strands of DNA attached to the flowcell. This creates “bridges” of double-stranded DNA between the primers on the flowcell surface. The double-stranded DNA is then broken down into single-stranded DNA using heat, leaving several million dense clusters of identical DNA sequences. When cluster generation is complete, the templates are ready for sequencing.
- Sequencing: Illumina® uses a proprietary reversible terminator based method that detects single bases as they are incorporated into DNA template strands. Primers and fluorescently labeled terminators (terminators are a version of nucleotide base (A, C, G or T) that stop DNA synthesis) are added to the flow cell. The primer attaches to the DNA being sequenced. The DNA polymerase then binds to the primer and adds the first fluorescently labelled terminator to the new DNA strand. Once a base has been added no more bases can be added to the strand of DNA until the terminator base is cut from the DNA. Each of the terminator bases (A, C, G, and T) give off a different colour. The fluorescently labelled terminator group is then removed from the first base and the next fluorescently base can be beside added. And so, the process continues until millions of clusters have been sequenced.
- Data Analysis: During data analysis and alignment, the newly identified sequence reads are aligned to a reference genome, this looks for matches or changes in the sequenced DNA. Following alignment, many variations of analysis are possible: DNA sequencing may be used to determine the sequence of individual genes, larger genetic regions (clusters of genes), full chromosomes, or entire genomes.
For a detailed animation of Illumina® sequencing technology click and watch this video -->
2. Paired-end sequencing [edit]
(Erminda Ferro)
All Illumina next-generation sequencing (NGS) systems are capable of paired-end sequencing : there are different steps ffor end pair sequencing, as you can see in the last part of the video (
- the forward sequence is transcribed into the read1.
- the read 1 is washed away
- the farward sequence is transcribed into a reverse sequence
- the farward sequence is washed away
- the remaining reverse sequence is the template for the read2.
With this technique you obtain both the reverse (read2) and the forward (read1) sequencing of each piece of sequence.
In general, regulating the number of the cycles, it is possible to determine the length of the reads and having or not overlapping read1 and read2.
There are different advantages for this technique, for example:
- Efficient Sample Use: Requires the same amount of DNA as single-read genomic DNA or cDNA sequencing
- Simple Data Analysis: Enables high-quality sequence assemblies with short-insert libraries. A simple modification to the standard single-read library preparation process facilitates reading both the forward and reverse template strands of each cluster during one paired-end read. Both reads contain long-range positional information, allowing for highly precise alignment of reads
So paired-read sequencing produces twice the number of reads for the same time and effort in library preparation and enables more accurate read alignment.
In particular, if we are talking about DNA seq , this technique facilitates detection of genomic rearrangement, such as indel and inversions, and repetitive sequence elements(fig below) .
If we perform RNA seq we can more easily detect genes fusion and novel splice isoforms.

3. Ion torrent sequencing [edit]
(Matteo Vayr)
Ion torrent sequencing is a method of DNA sequencing based on the release of H+ ions as byproducts of nucleotides incorporation.
![]()
An array of microwells on a semiconductor chip present beads with different template DNA strands and DNA polymerase. These microwelles are flooded with a solution containing single species of deoxyribonucleotide triphosphate (dNTP; A C G or T). If the dNTP is incorporated by the DNA polymerase on the tamplate strand, an H+ ion is released, this leads to a current change in the solution that is detected by an ion sensor present in each microwell. The activation of the sensor is registered and the corresponding nucleotide added to the sequence.
If two dNTPs of the same type are incorporated, 2 ions are released and the current change is double, than 2 nucleotide (e.g. TT) are added to the sequence.

After that the dNTP solution is washed away and another solution with another type of dNTP is added in the microwells and so on until the complete sequencing.
If no dNTP is integrated no current change will occour and it will be simply washed away without adding it to the sequence and the microwell flooded again with another dNTP.
This technique is different from others because based on a binary system (current or no current) and work without using modified nucleotides or optic changes, furthermore is cheaper and faster (100-200 nucleotides per hour) than others sequencing techniques.
4. Roche sequencing [edit]
(Maria Giovanna Caruso)
Roche sequencing is another technology of NGS; it’s able to work on genomic DNA, DNA produced by PCR and cDNA.
Sequence length
of 400 – 1000 bases are read, so, at the starting point it is necessary to
reduce the sequence, thanks to a nebulizer, in smaller fragments (fragmentation). After creating single
strand fragments, it’s necessary to bind two different adaptors (for example A
and B) at the two ends (adaptors ligation):
one of the adaptors helps the DNA fragment to attack the beads (insoluble small
particles or nano spheres), since it is complementary to the sequence attacked
to the bead. So, small single strand fragments of DNA will surround the beads. Then, there is the amplification of the
target to get better and accurate sequencing results: to do this, there is
firstly the production of chains starting from the single strand fragments, and
then, these complementary sequences will be used for the amplification (having terminal
sequences complementary to the primers). Now, the beads can be picked and loaded
in specific wells to be inserted in the sequencing machine (loading).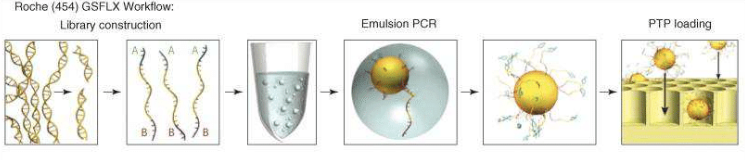
The next step is the pyrosequencing reaction: the main innovation of 454 Roche is the use of inorganic pyrophosphate instead of fluorescent nucleotides, to generate the fluorescence. PPi is the substrate of the enzyme sulfurylase that produce ATP from PPi and AMP. ATP is used by a luciferase that oxidises the luciferin, creating light signals. Nucleotides are added one by one to analyse the fluorescence deriving from the ones that have bound the target sequence.
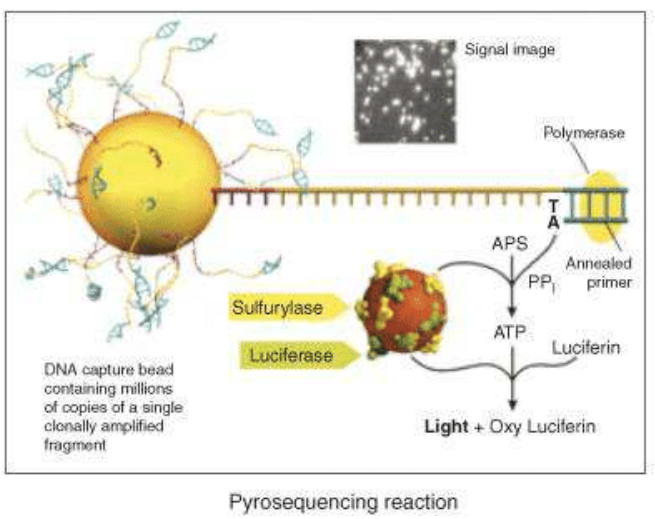
A photodetector can, finally, read the signals and produce a graph expressing the correct sequence. Roche technology currently uses two systems based on the pyrosequencing technology: the GS FLX+ and the GS Junior System. The GS FLX+ System promises read lengths of approximately 1000 base pairs while the GS Junior System promises 400 base pair reads (data analysis).
One of the advantages of 454 systems is their high running speed [sample/day 3840].
There are also some disadvantages:
· the 454 system is prone to errors when estimating the number of bases in a long string of identical nucleotides (homopolymer error);
· the price of reagents is relatively more expensive compared with other next-generation sequencers.
5. Pacific Bio and Nanopore long-run sequencing [edit]
(Benedetta Arlorio)
PacBio long-read sequencing
PacBio long-read sequencing enabled by SMRT Sequencing technology allows real-time observation of DNA synthesis and offers longer read lengths than the second-generation sequencing (SGS) technologies, making it well-suited for unsolved problems in genome, transcriptome, and epigenetics research.
video:
SMRT Sequencing is built upon two key innovations:
- zero-mode waveguides (ZMWs): allow light to illuminate only the bottom of a well in which a DNA polymerase/template complex is immobilized.
- Phospholinked nucleotides: allow observation of the immobilized complex as the DNA polymerase produces a completely natural DNA strand.
Each of the four nucleotides is labeled with a different fluorescent dye (indicated in red, yellow, green, and blue, respectively for G, C, T, and A) so that they have distinct emission spectrums. As a nucleotide is held in the detection volume by the polymerase, a light pulse is produced that identifies the base.

Operating mechanism:
- A fluorescently-labeled nucleotide associates with the template in the active site of the polymerase.
- The fluorescence output of the color corresponding to the incorporated base is elevated.
- The dye-linker-pyrophosphate product is cleaved from the nucleotide and diffuses out of the ZMW, ending the fluorescence pulse.
- The polymerase translocates to the next position.
- The next nucleotide associates with the template in the active site of the polymerase, initiating the next fluorescence pulse.
Advantages:
- longer average read lenghts: the PacBio RS II system with the current C4 chemistry boasts average read lengths over 10 kb and maximum read lengths over 60 kb (the maximum read length of Illumina HiSeq 2500 is only paired-end 250 bp).
- high consensus accuracy.
- uniform coverage: this technology does not require an amplification step, leading to uniform coverage across all genome: this allows you to sequence through palindromes and low-diversity regions of the genome. These long reads also allow for the spanning of complex regions.
- simultaneous epigenetic characterization: with this technology you can automatically detect DNA modifications and measure the rate of DNA base incorporation during sequencing: DNA polymerization runs freely at ~3 bases/second. Alteration of this rate due to the incorporation of nucleotides across modified bases is detected and used to infer the presence of bases other than A, C, T or G. This information is automatically generated and processed during every run.
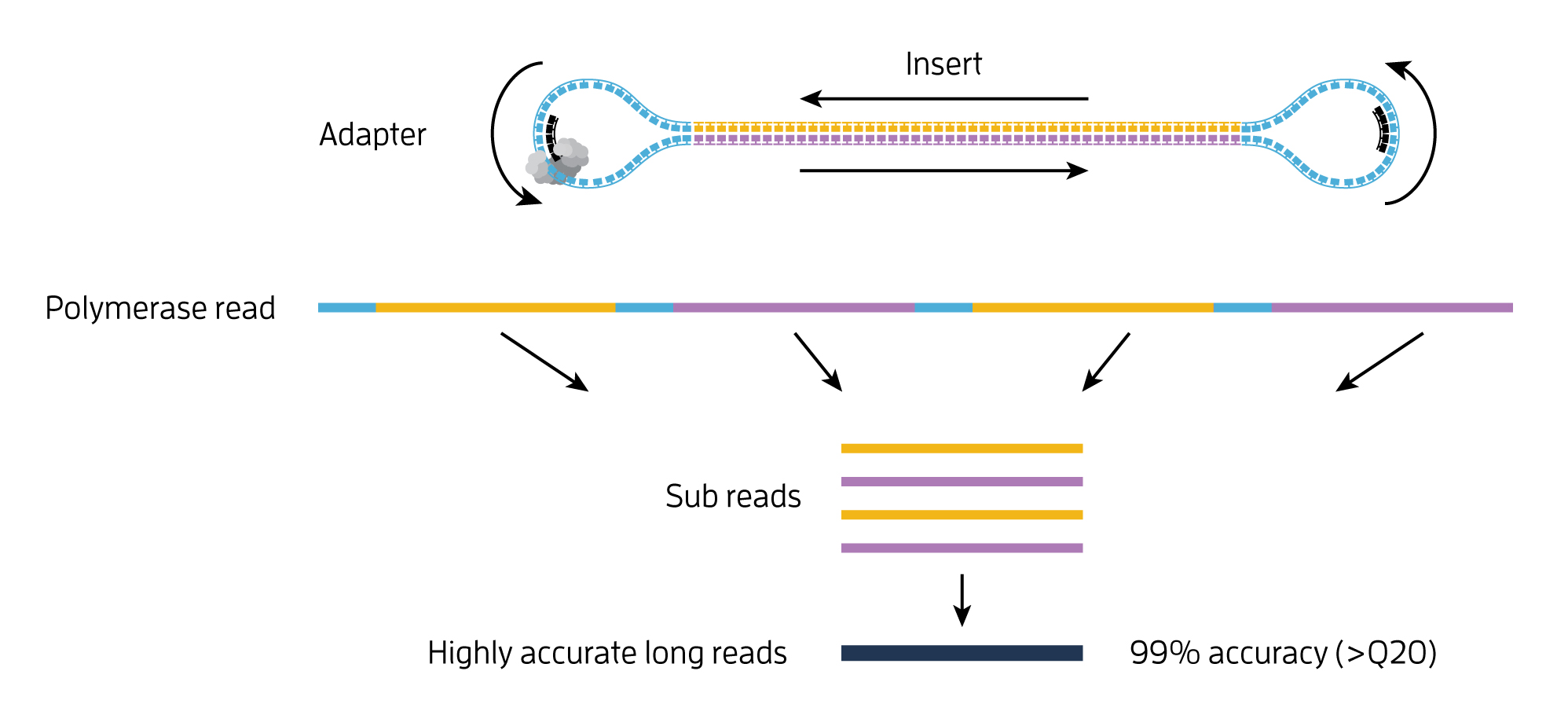
- single-molecule resolution: Single Molecule, Real-Time (SMRT) Sequencing uses the Circular Consensus Sequencing (CCS) method to generate highly accurate long reads with >99% single-molecule read accuracy. The CCS method generates a highly accurate reads (HiFi Reads) based on building consensus from the multiple passes of forward and reverse strands of the insert DNA incorporated into a SMRTbell molecule.
Nanopore sequencing
A nanopore is a nano-scale hole through whcich passes an ionic current and we can measure the changes in current as biological molecules pass through the nanopore or near it. The information about the change in current can be used to identify that molecule.
Holes can be created by proteins puncturing membranes (biological nanopores) or in solid materials (solid-state nanopores). Protein nanopores can be adapted at Angstrom-level precision using protein-engineering techniques. Specific adaptations can be designed so that the nanopore is a sensor for a range of specific molecules.
video:
Operating mechanism:
A protein nanopore is set in an electrically resistant polymer membrane. An ionic current is passed through the nanopore by setting a voltage across this membrane. A DNA strand is passed through the pore: this event creates a characteristic disruption in current (as shown in the diagram below). As a strand of DNA emerges from a nanopore, a `phosphate grabber' on one functionalized electrode and a `base reader' on the other electrode form hydrogen bonds to complete a transverse electrical circuit through each nucleotide as it is translocated through the nanopore. Measurement of that current makes it possible to identify the molecule in question.
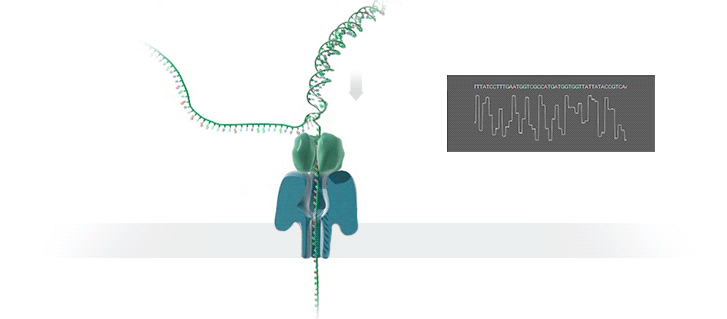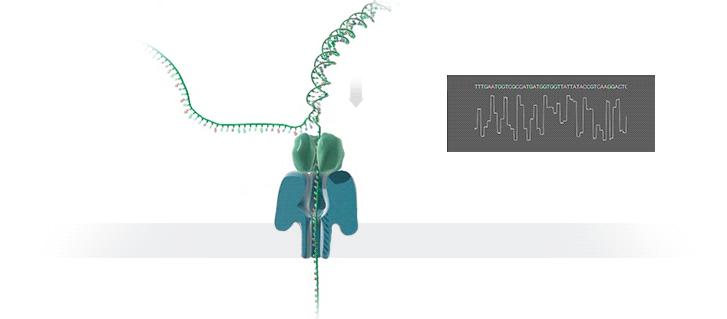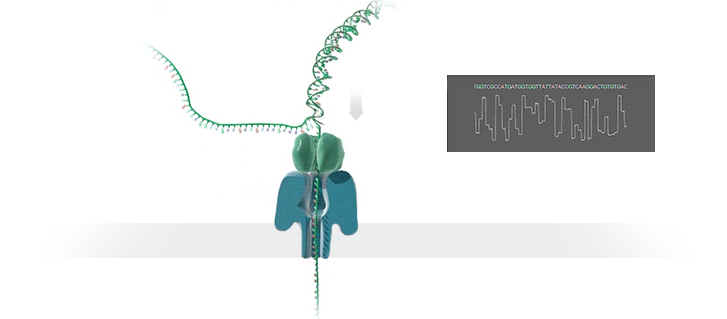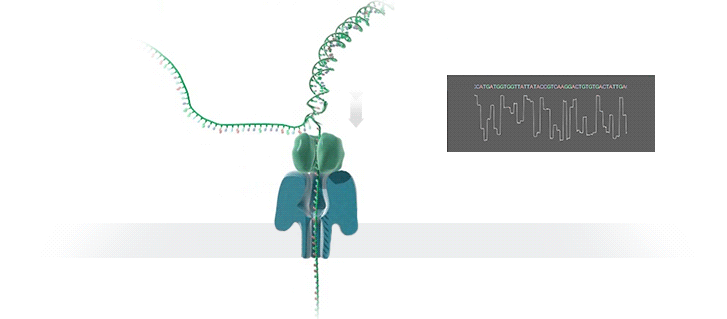
Types of nanopore:
- Biological: relies on the use of transmembrane proteins, called porins, that are embedded in lipid membranes so as to create size dependent porous surfaces- with nanometer scale "holes" distributed across the membranes. Sufficiently low translocation velocity can be attained through the incorporation of various proteins that facilitate the movement of DNA or RNA through the pores of the lipid membranes.
- alpha emolysin: The structure of αHL is advantageous to identify specific bases moving through the pore. The αHL pore is ~10 nm long, with two distinct 5 nm sections. The upper section consists of a larger, vestibule-like structure and the lower section consists of three possible recognition sites (R1, R2, R3), and is able to discriminate between each base.
- Mycobacterium smegmatis porin A (MspA): a natural MspA, while favorable for DNA sequencing because of shape and diameter, has a negative core that prohibited single stranded DNA(ssDNA) translocation. The natural nanopore was modified to improve translocation by replacing three negatively charged aspartic acids with neutral asparagines. The electric current detection of nucleotides across the membrane has been shown to be tenfold more specific than αHL for identifying bases.
- solid state: Solid state nanopore sequencing approaches, unlike biological nanopore sequencing, do not incorporate proteins into their systems. Instead, this technology uses various metal or metal alloy substrates with nanometer sized pores that allow DNA or RNA to pass through. These substrates most often serve integral roles in the sequence recognition of nucleic acids as they translocate through the channels along the substrates.
An effective technique to determine a DNA sequence has been developed using solid state nanopores and fluorescence. This fluorescence sequencing method converts each base into a characteristic representation of multiple nucleotides which bind to a fluorescent probe strand-forming dsDNA. With the two color system proposed, each base is identified by two separate fluorescences, and will therefore be converted into two specific sequences. Probes consist of a fluorophore and quencher at the start and end of each sequence, respectively. Each fluorophore will be extinguished by the quencher at the end of the preceding sequence. When the dsDNA is translocating through a solid state nanopore, the probe strand will be stripped off, and the upstream fluorophore will fluoresce. This sequencing method has a capacity of 50-250 bases per second per pore, and a four color fluorophore system (each base could be converted to one sequence instead of two), will sequence over 500 bases per second. Advantages of this method are based on the clear sequencing readout—using a camera instead of noisy current methods. However, the method does require sample preparation to convert each base into an expanded binary code before sequencing. Instead of one base being identified as it translocates through the pore, ~12 bases are required to find the sequence of one base.
Advantages:
- minimal sample preparation: would not require the use of purified fluorescent reagents and would use unamplified genomic DNA, thus eliminating enzymes, cloning and amplification steps.
- sequence readout that does not require nucleotides, polymerases or ligases.
- very long read-lengths (>10,000-50,000 nt).
- very inexpensive
Challenges:
none of the natural or manmade nanopore structures used to date has had the appropriate geometry to detect the features of only one nucleotide at a time while the polymer is translocating through the pore: none of these nanopores has channels shorter than ~5 nm and, because at least 10-15 nucleotides of ssDNA extend through a channel of this length, all of these nucleotides together contribute to the ionic current blockade. so, at this stage of the technology, a substantial short-term challenge is to slow DNA translocation from microseconds per base to milliseconds, and several recent studies indicate that this can be achieved by using DNA-processing enzymes. If a future instrument incorporates the hemolysin heptamer, it will also be necessary to establish a stable support of some kind. Again, there is recent progress toward this end, though in the longer term it seems likely that synthetic solid-state nanopores will be preferred for a commercial instrument. Electronic sensing based on either tunneling probes or a capacitor is being tested for its ability to detect a DNA strand during translocation, but whether this is possible remains to be demonstrated. A continuing concern is that stochastic motion of the DNA molecule in transit will increase signal noise in such a sensor, thereby reducing the potential for single-base resolution.
