Knocking-down and knocking-out genes
(Restore this version)
Modified: 17 April 2019, 3:55 PM User: Ilaria Ghia →
Back to index
Tentative subjects:
1. Transient siRNA/LNA-mediated knock-down
2. Stable shRNA knock-down
3. K.O. in cultured cells, alternatives
(Ilaria Ghia)
A knockout cell line (constitutive or conventional KO) is a model in which a target gene is permanently inactivated and thus is not able to produce the protein it encodes for.
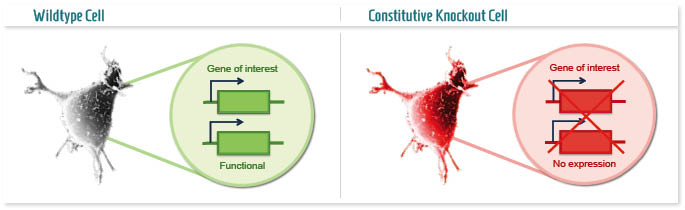
The applications of this technology are many, and can be distinguished on the type of research it is used for:
- Academic research: study of the main functions of a gene and/or protein, LOF (Loss-of-function) studies, in vitro recapitulation of human diseases;
- Bio-pharmaceutical research: in vitro target validation, drug study and screening through mimicry of human diseases,study of safety, off-target activity and specificity of a drug.
As every technique, knockout cell lines show some strength and limitations. Three main advantages are:
- Total absence of protein, excluding all the possible isoforms;
- Since each cell line is matched with an isogenic (=genetically identical) control cell line, it is a reliable methodology;
- Feasible in every genetic background or cell type.
By contrast, three weaknesses are:
- Genetic compensation: the function of the KO protein can be supplied by the action of other related proteins;
- Non-specific phenotype: the editing of a gene can sometimes affect the neighboring genes too;
- Not very suitable for highly polyploid cell lines.
Nevertheless, these limitations can be minimized with bioinformatic, genetic and bibliographic analyses. Moreover, an alternative is represented by functional knockouts, in which the use of CRISPR/Cas9 or other genome editing methods allows the introduction of a point mutation to produce an inactive protein.
The generation of KO cell lines requires several steps, summarized by the following image.

Clone screening is needed to select the clones that correctly integrated the plasmid, which come in the end in a small number, reducing the efficiency of the process. An alternative is to use the newest technologies that involve some enzymes, such as TALEN and ZNF (besides the most diffused CRISPR/Cas9, which is discussed in the Genome Editing section of this Wiki: http://cmb.i-learn.unito.it/mod/wiki/view.php?pageid=114).
