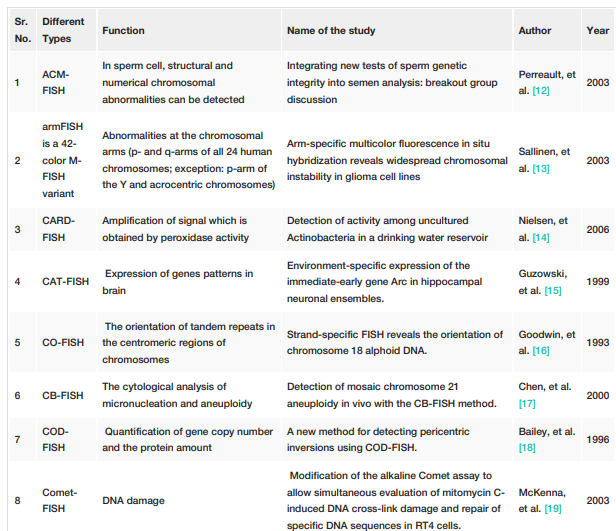
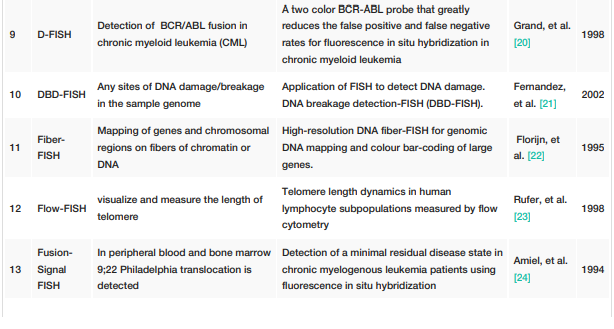
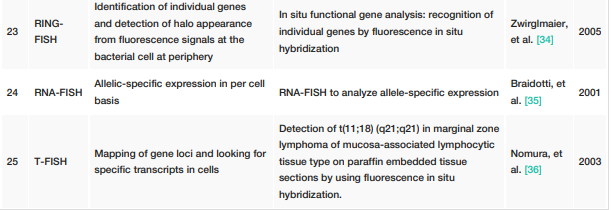
FISH techniques
(Restore this version)
Modified: 28 April 2019, 6:57 PM User: Katie Znidericz →
Tentative paragraphs:
1. Introduction
2. RNA-FISH
3. Immuno-FISH
4. Chromosome painting
5. FISH: advantages and disadvantages
1. Introduction
(Simone Rocco): The development of banding techniques in the 70’s led to a first revolution in cytogenetic field, allowing an accurate definition of chromosomes and their abnormalities. The karyotyping study has increased our knowledge of the onco-hematology field, becoming necessary for a correct diagnostic classification. The major limitation of conventional cytogenetics is that the cell must be in mitosis in order to be correctly analyzed. Moreover, despite the presence of high resolution methods, the conventional cytogenetic analysis is able to detect only chromosomal rearrangements that affect more than 3 megabases (Mb). All these problems have been resolved thanks to the introduction of the Flourescence in situ hybridization (FISH) at the end of the 80’s.
(Rachele Rosso): FISH is a kind of cytogenetic technique which uses fluorescent probes binding parts of the chromosome to show a high degree of sequence complementarity, and it can be used to find out where the fluorescent probe bound to the chromosome. FISH is often used for finding specific features in DNA, and to detect and localize specific RNA targets (mRNA, lncRNA and miRNA).This technique provides a novel way for researchers to visualize and map the genetic material in an individual cell, including specific genes or portions of genes. Different from most other techniques used for chromosomes study, FISH has no need to be performed on cells that are actively dividing, which makes it a very versatile procedure. It is composed by 4 main passages:
- A probe complementary to the known sequence is made and it is labelled with a fluorescent marker, as for example fluorescein, by incorporating nucleotides that have the marker attached to them;
- Chromosomes are put on a microscope slide and denatured;
- The probe is denatured and added to the microscope slide, allowing the probe hybridize to its complementary site;
- The excess probe is washed off and the chromosomes are observed under a fluorescent microscope. The probe will show as one or more fluorescent signals in the microscope, depending on how many sites it can hybridize to.

FISH is widely used for several diagnostic applications as for example identification of numerical and structural abnormalities, characterization of marker chromosomes, monitoring the effects of therapy, detection of minimal residual disease. Moreover it has many applications in research such as gene mapping or identification of amplified genes. FISH is also used to compare the genomes of two biological species to deduce evolutionary relationships.
2. RNA-FISH
(Linda Petrucci): RNA Fluorescence in Situ Hybridization is a specific type of in situ hybridization targeting RNA instead of DNA molecules using probes complementary to the target RNA. These probes can be detected with fluorescence microscopy by either direct detection or using immunofluorescent amplification techniques.
Direct detection is possible by using a set of probes, usually single stranded DNA oligonucleotides, labelled with one or more fluorescent dyes in order to have enough visibility to overstep the background autofluorenscence and thus to be detected with a specific microscope. The use of a set of probes instead of a single one enhance the possibility to distinguish the probe itself to the real RNA, much brighter, reducing numbers of false positives. False negatives are really unlikely because the chances that all the probes fail to bind is pretty low. Direct detection gives also the possibility to use different colours for different target at the same time, in order to distinguish them from the others during the analysis.
Indirect techniques actually avoid all the problems due to background fluorescence in the direct detection, indeed lots of methods to amplify the probe signal were found, including indirect immunochemistry or binding happens to the probes that can be recognized by antibodies conjugated to enzymes, e.g. biotin-streptavidin reaction. The only problem with this method is the high numbers of steps that increase the possibility of error and the general cost of the technique.
RNA fish is mostly used to detect gene expression, especially in developmental biology. One of the main advantages offered by using the FISH technology is that it makes it possible for us to follow the target’s movements inside a living cell, allowing researcher to study, for example, gene disposition in the nucleus, and to highlight the main differences between different cell lines set of genes’ expression. This technique allows RNA detection in case of both symmetrical cellular distribution, and asymmetrical subcellular distribution of RNAs.
3. Immuno-FISH(Katie Znidericz): Over the years the FISH technique has been adapted and diversified in order to achieve a wider ranged diagnostic tool with improved capabilities such as the improvement in sensitivity, specificity and resolution. One of these adaptations is Immuno-FISH.
As the name suggests it is a combination of two valuable techniques - immunostaining and FISH, in order for us to analyse the given sample not only from the point of view of the genotype but also the phenotype ie. the proteins that are expressed. It is easy to perform and no more time consuming than the usual FISH technique. This method allows us to visualise of chromosome territories, chromosome subregions, single genes, and RNA transcripts preserving their spatial positions in the cell nucleus.
Immuno-FISH can be used both with the 2D flattened chromosome and the 3D preserved nuclei, and it is with the 3D FISH that we can actually analyse both the DNA and the proteins in the same sample. However some steps of the FISH procedure may interfere with immunostaining. The “new generation” of confocal microscopes that allow the distinct visualizations of at least five different fluorochromes within one experiment opened the way for multicolor 3D-FISH experiments. And so, many differently labeled nuclear targets can be localised simultaneously.
While FISH can be combined with the detection of cellular and nuclear proteins, there may arise some issues with detection following permeabilisation pretreatment and DNA denaturation, as some antigens may not be stable enough to withstand them. That is why it is important when planning the experiment to use different strategies depending on the protein for example it may be necessary to use primary and secondary antibodies.
Usually protocols suggest fixation, pretreatments, hybridization before the FISH technique. However an efficient hybridization requires a number of permeabilization steps and the denaturation of cell DNA. These steps have to be carefully balanced, in order to maintain the best possible nuclear morphology on one hand and making chromatin accessible for probe penetration on the other hand. Small deviations or experimental mistakes can easily change the quality of the experimental outcome.
4. Chromosome painting
(Laura Zanatto): Chromosome painting refers to the hybridization of fluorescently labeled chromosome-specific, composite probe pools to cytological preparations. With FISH, chromosome painting can be used to analyze the entire genome, allowing one to screen for chromosomal aberrations. Before FISH, cytogenetic screening tests for numerical and structural aberrations were restricted to conventional chromosome banding analyses. Chromosome painting, however, has also become a versatile tool in basic research disciplines ranging from radiation biology, to evolutionary cytogenetics, and research dealing with aspects of the nuclear structure.
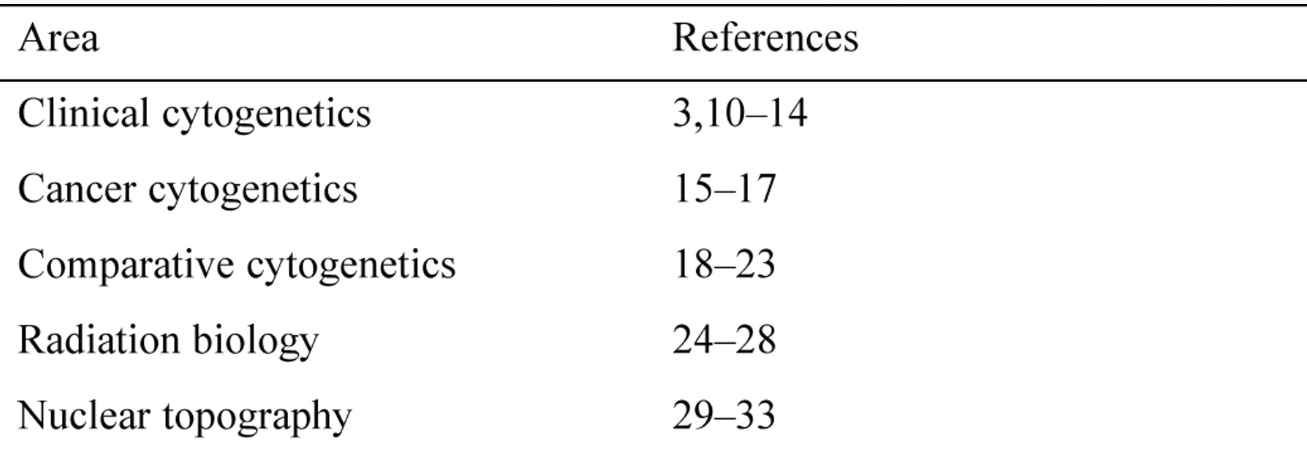
Chromosome painting probes have been improved rapidly and modified in several aspects. The first generation of probes, based on chromosome-specific phage libraries, were rather cumbersome to use, due to low insert-to-vector ratios which frequently resulted in a relatively high background staining. Some of these limitations were overcome with the availability of plasmid libraries where an improved insert-to-vector ratio and easier probe generation enhanced the painting quality considerably. Two alternative approaches were used to reach the goal of color karyotyping chromosomes:
1. Fluorochrome-specific optical filters, termed m-FISH;
2. Interferometer-based spectral imaging (introduced as spectral karyotyping or SKY).
(Rachele Rosso): The multiplex in situ hybridization (M-FISH) represents one of the most significant developments in molecular cytogenetics of the past decade; it is a 24-color karyotyping technique and is the method of choice for studying complex interchromosomal earrangements. The process is composed by three main steps:
- The multiplex labeling of all chromosomes in the genome with finite numbers of spectrally distinct fluorophores such that each homologous pair of chromosomes is uniquely labelled;
- The microscopic visualization and digital acquisition of each fluorophore using specific single band-pass filter sets and dedicated M-FISH software. These acquired images are then superimposed enabling individual chromosomes to be classified based on the fluor composition in accordance with the combinatorial labeling scheme of the M-FISH probe cocktail used;
- The detailed analysis of these digitally acquired and processed images to resolve structural and numerical abnormalities.
(Laura Zanatto): Spectral karyotyping (SKY) is a FISH technique that refers to the molecular cytogenetic analysis of metaphase preparations by means of spectral microscopy. Screening cell karyotypes for chromosomal abnormalities is an integral part of the diagnosis of human cancers and congenital diseases and SKY allows to paint each of the 24 human chromosomes with different colours. SKY is based on a single exposure through a triple-bandpass optical filter and spectroscopic analysis. Spectral imaging is not dependent on fluorescence intensity but solely on spectral signatures created by combinatorial labelling.
After metaphase preparations, probes are hybridized;
chromosome-specific painting probes are obtained by flow sorting and amplified
by two rounds of DOP-PCR. Then there is the denaturation of both sample and
probes, and following hybridization. After in situ hybridisation and
immunodetection, a spectral image is acquired using a conventional fluorescence
light microscope.
SKY will be very useful in clinical cytogenetics and allow us to identify traslocation, deletion or alteration in metaphasic chromosomes, but it is not possible to evaluate structural abnormalities, such as inversion, deletion, insertion, and duplication in the same chromosome, because these are shown with the same color.
SKY will be very helpful for cytogenetic analysis of
solid tumours, because cancer cell karyotypes can be very complex, with a lot
of aberrations. Some probe sets are based on repetitive centromeric
satellite Probes, that can be used for some diagnosis. Also haematological
malignancies can be studied with FISH, such as some cases of leukaemia and
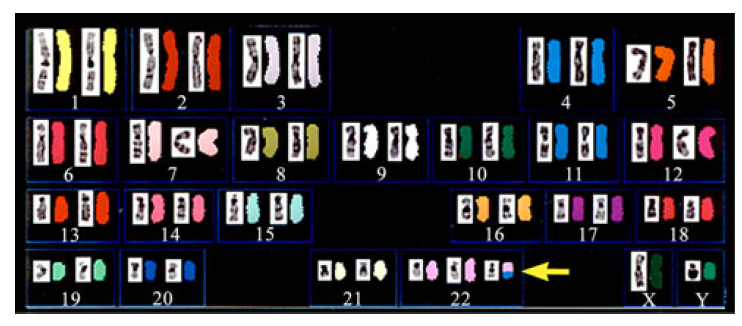
lymphoma (Fig: AML leukaemia). The results of chromosome
analysis on Trisomy 11/22 using SKY are shown in Fig: as we can see SKY is
one of the most effective approaches at present for the identification of an
extra chromosome of unknown origin. This technique can be also used in
comparative cytogenetic to define chromosomal
rearrangements that occurred during the course of evolution.

5. FISH: advantages and disadvantages
(Simone Rocco): This technique is based on the ability of a DNA probe, marked with a fluorophore, to bind specifically to a complementary DNA target sequence. FISH can be used for metaphasic chromosomes, interphase nuclei, chromatin fibers or DNA microarrays. An interesting point is that the FISH conducted on interphase cells (Interphase FISH or iFISH) was the method that produced the greatest results in onco-hematology diagnostics. The probes used in FISH can be different: the whole chromosome painting probes permitted us to indentify chromosomes involved in structural anomalies, while the single-locus probes were fundamental not only in gene mapping, but also in the detection of break points in chromosomal translocations.
The chronic lymphatic leukemia (CLL) is the best example to understand what was the contribution of iFISH in the study of cell populations with a mitotic index of less than 1%. The chromosome analysis of patients suffering from CLL often does not provide results for the absence of metaphase states or demonstrates a normal chromosomal kit in 40-50% of cases. Instead, iFISH identifies abnormalities of the karyotype in 65% of patients analyzed during the diagnosis or during a stable phase of the disease and in 88% of patients with a progressive disease (?). Another example is the acute B-cell lymphoblastic leukemia (ALL): 106 karyotype alterations were defined thanks to FISH, while only 34 with banding techniques.
However, we have to consider that this technique has its limits. In fact, we must know in advance what type on chromosomal anomaly we expect to find, in order to be able to choose the most appropriate probe for the analysis. Furthermore, since a limited number of fluorochromes can be used, the FISH is not able to analyze simultaneously more than three anomalies. However, also all these limits have been recently resolved thanks to the introduction of multiplex-FISH (M-FISH) and Spectral Karyotyping (SKY), which were previously discussed. In terms of using the FISH technology around the world, we found that the FISH technique has very limited use in developing countries because of unavailability and lack of expert knowledge.
Other types of FISH: