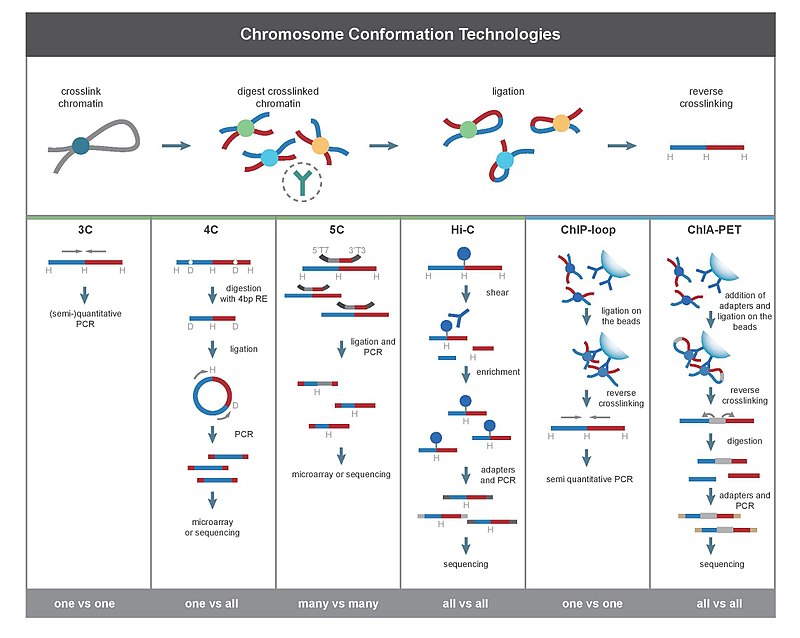
Long-range interaction and chromatin loops: from 3C to Hi-C
Back to index
(Mirko Scrivano)
3C technology (Chromosome Conformation Capture): technology used to identify whether two known genomic regions interact (one vs one) . After the crosslinking of the two strands (formaldehyde), a digestion with a common restriction enzyme is performed (usually EcoRI, whose site is present once every 400 bases, as mean). Than, intramolecular ligation is carried out, using Ligase as ATP (MILD CONDITION, to avoid intermolecular ligation). Now a reverse crosslinking step is performed. A phenol-clorophorm purification should be done, to eliminate proteins present in the mixture (*). Now, with specific primers (pairing the two regions of interest) is possible to detect the interaction by simple PCR.
4C-seq (Circularized Chromosome Conformation Capture): based on 3C technique, allows to identify the interaction(s) between a known genomic region and all other possible regions (one vs all). The first steps are the same of the 3C, until “*”. Now, a second digestion is performed, using likely a 4 bases cutting enzyme. The sticky ends produced will be then ligated together (Ligase and ATP) always under mild condition, forming a circular structure. Using primers of the known region a reverse PCR will be performed. The PCR products (library) will then be NGS sequenced to identify the interacting region(s).
(Edoardo Alfì) 5C and Hi-C
5C (Chromosome Conformation Capture Carbon Copy) detects interactions between all restriction fragments within a given region (typically no greater than a megabase). This is done by ligating universal primers to all fragments. However, 5C has relatively low coverage. The 5C technique overcomes the junctional problems at the intramolecular ligation step and is useful for constructing complex interactions of specific loci of interest. This approach is unsuitable for conducting genome-wide complex interactions since that will require millions of 5C primers to be used.
Here, a short scheme to resume

Hi-C uses high-throughput sequencing to find the nucleotide sequence of fragments. The original protocol used paired end sequencing, which retrieves a short sequence from each end of each ligated fragment. As such, for a given ligated fragment, the two sequences obtained should represent two different restriction fragments that were ligated together in the random ligation step. The pair of sequences are individually aligned to the genome, thus determining the fragments involved in that ligation event. Hence, all possible pairwise interactions between fragments are tested.
Hi-C assay:
(Noemi Dorma) ChIA-PET(Chromatin Interaction Analysis by Paired-End Tag Sequencing)
It's a technique that combines 3C with ChIP-seq but, differently from ChIP loop, allows the identification of all the possible interacting loci , whose interaction is mediated by a specific transcription factor, the target of the IP (all vs all). After formaldehyde crossilink and sonication , IP of the protein of interest is performed (enrichment). Byotinilated adapters are added at the ends of the fragmented DNA and subsequent intramolecular ligation is performed . Then, another restriction enzyme cut is performed and other adapters are added at the 5'/3' protruding ends. PCR amplification and NGS sequencing are finally performed.
(Alessandro Ferrando) ChIP-loop
Last but not least, this technique, that combines 3C (two precise loci) with ChIP-seq, is used to identify whether an interaction occurs between two selected loci, mediated by a specific protein (one vs one). This technique increases the specificity of 3C by selecting for a known protein mediating the DNA-DNA interaction.
To sum up...