Knocking-down and knocking-out genes
Back to index
Tentative subjects:
1. Transient siRNA/LNA-mediated knock-down
2. Stable shRNA knock-down
3. K.O. in cultured cells, alternatives
(Sara Barsanti)
Transient knockdown is the change in gene expression caused by an oligonucleotide binding to an mRNA or temporarily binding to a gene. This leads to a temporary change in gene expression that does not modify the chromosomal DNA. The most direct use of transient knockdowns is for learning about a gene that has been sequenced, but has an unknown or incompletely known function.
RNA interference (RNAi) is a means of silencing genes by way of mRNA degradation. Gene knockdown by this method is achieved by introducing small double-stranded interfering RNAs (siRNA) (a class of double-stranded RNA molecule, 20-25 base pairs in length) into the cytoplasm. Small interfering RNAs can originate from inside the cell or can be exogenously introduced into the cell. Once introduced into the cell, exogenous siRNAs are processed by the RNA-induced silencing complex (RISC). The siRNA is complementary to the target mRNA to be silenced, and the RISC uses the siRNA as a template for locating the target mRNA. After the RISC localizes to the target mRNA, the RNA is cleaved by a ribonuclease.
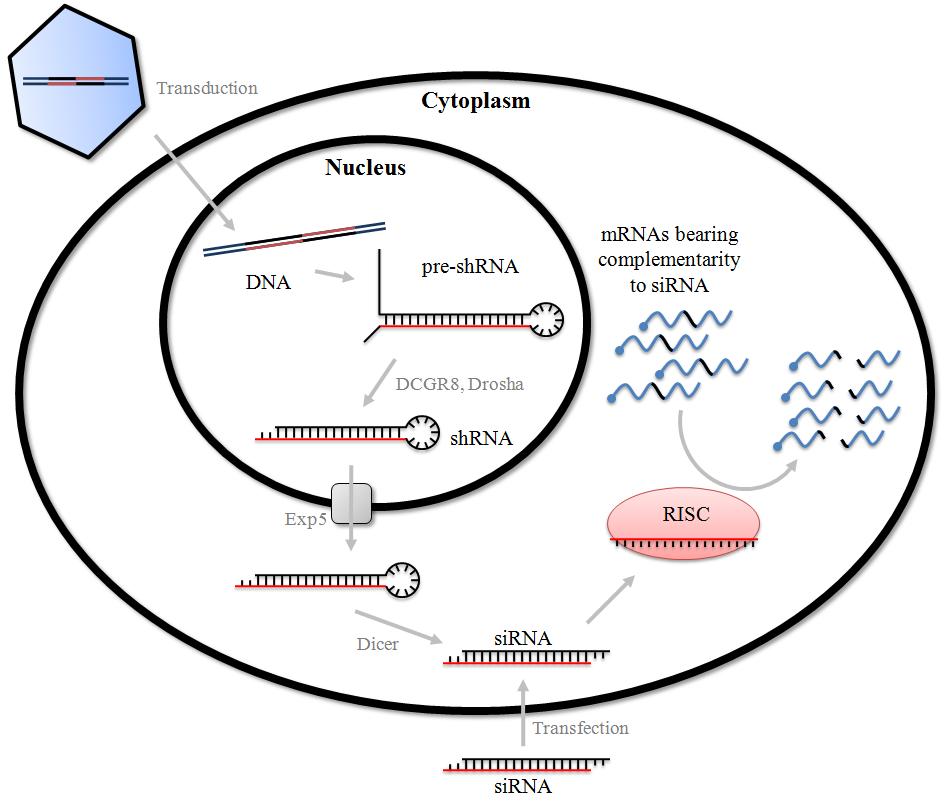
Because RNAi intersects with a number of other pathways, it is not surprising that on occasion nonspecific effects are triggered by the experimental introduction of an siRNA. When a mammalian cell encounters a double-stranded RNA such as an siRNA, it may mistake it as a viral by-product and mount an immune response. Furthermore, because structurally related microRNA modulate gene expression largely via incomplete complementarity base pair interactions with a target mRNA, the introduction of an siRNA may cause unintended off-targeting.
A locked nucleic acid (LNA), often referred to as inaccessible RNA, is a modified RNA nucleotide in which the ribose moiety is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'-endo (North) conformation. LNA bases form standard Watson–Crick base pairs but increase the rate and stability of the basepairing reaction. LNAs also have increased affinity to base pair with RNA as compared with DNA. These properties render LNAs especially useful as probes for fluorescence in situ hybridization and comparative genomic hybridization, as antagonists for miRNAs, and as antisense oligonucleotides to block mRNA translation.
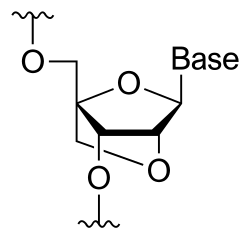
LNA designs can be divided in two main categories: mixmers and gapmers. In a mixmer, LNA and DNA are interspersed throughout the sequence of the oligonucleotide, whereas, in a gapmer, two LNA segments at both ends of the oligonucleotide are separated by a central segment or gap of DNA nucleosides.
To inhibit mRNA expression and protein translation gapmer designs are the most potent. This is because the central DNA/PS segment, which is longer than 7–8 DNA nucleotides (nt), recruits the RNA-cleaving enzyme RNase H when the gapmer is hybridized to the mRNA. Gapmers targeting both exons and introns work equally efficiently and, thus, all stages of mRNA and also ncRNA processing can be addressed. Mixmers can also be used to block translation, but must be designed to bind close to, at, or upstream of the translational start site to do so . The mixmer will inhibit translation by blocking the binding of the ribosomal subunits to the mRNA. Alternatively, mixmers binding close to the 5′ end of the pre-mRNA can prevent 5′-cap formation and thereby inhibit translation.

Inhibition, or sequestering, of miRNA is a gain of function that has attracted a special interest, miRNA are an important class of regulatory ncRNAs that can bind to partially complementary sites located in the 3′ untranslated regions (UTRs) of target mRNAs, promoting translational repression or deadenylation and degradation. Sequestering miRNAs by LNA mixmer hybridization has wide therapeutic potential.
(Giulia Pirro)
The RNAi (RNA interference) is a class of dsRNA that allows the silencing of a target gene.
The shRNA (short hairpin RNA or small hairpin RNA) is an artificial construct which is able to bind in a specific way the mRNA coding for the known gene.
The sequence of the shRNA is formed by a sense strand, an antisense strand (able to anneal with the previous) and a loop region in between.

Usually, shRNA is the key in order to obtain a knock-down. If the gene targeted for knock-down has a known biological or physiological function, would be easier and be extremely useful in testing the efficacy of an shRNA.
STEPS IN ORDER TO SILENCE A TARGET GENE:
- First of all the plasmids, which contains the shRNA construct, should be transfected in the cells. There are several ways in order to obtain the transfection, one of the most common is the lipid-base transfection.
- If these transfections occur, each cell is able to express the shRNA.
- The hairpin loop, which strongly characterize the molecule, has to be removed by the DICER. After this processing the shRNA will become siRNA.
- SiRNA is able to bind the protein complex RISC (RNA-Induced Silencing Complex) and from dsRNA (double-stranded RNA) it becomes single strand RNA.
- The complex formed by both RISC and the single stranded RNA, is able to bind the mRNA target, which is complementary to the shRNA construct, and cleaves it.
The crucial point between these steps is the stable transfection. The plasmid should contain both the shRNA construct but also a resistance gene. In this way is easier both the selection and the isolation of successfully transfected cell.

The crucial point between these steps is the stable transfection. The plasmid should contain both the shRNA construct but also a resistance gene. In this way is easier both the selection and the isolation of successfully transfected cell.
In order to better understand if the actually phenotype is the result of this treatment (and exclude any unspecific effect) is necessary perform few proves:- REDUNDANCY: producing more than one shRNA construct. These constructs share the same translation, but not the same nucleotides sequence. If these constructs are specific for a target mRNA the results will be the same.
- SCRAMBLED: produce the shRNA construct which doesn’t show a perfect matching. With-out the match should be express the target gene. Silencing doesn’t occur. Usually this mechanism is used like a negative control.
This kind of technique is largely use in biotechnology field in order to obtain a specific pattern of expres-sion and moreover looking forward a specific link between the expression and the associated phenotype.
(Ilaria Ghia)
A knockout cell line (constitutive or conventional KO) is a model in which a target gene is permanently inactivated and thus is not able to produce the protein it encodes for.
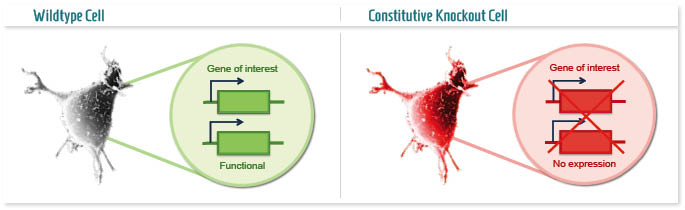
The applications of this technology are many, and can be distinguished on the type of research it is used for:
- Academic research: study of the main functions of a gene and/or protein, LOF (Loss-of-function) studies, in vitro recapitulation of human diseases;
- Bio-pharmaceutical research: in vitro target validation, drug study and screening through mimicry of human diseases,study of safety, off-target activity and specificity of a drug.
As every technique, knockout cell lines show some strength and limitations. Three main advantages are:
- Total absence of protein, excluding all the possible isoforms;
- Since each cell line is matched with an isogenic (=genetically identical) control cell line, it is a reliable methodology;
- Feasible in every genetic background or cell type.
By contrast, three weaknesses are:
- Genetic compensation: the function of the KO protein can be supplied by the action of other related proteins;
- Non-specific phenotype: the editing of a gene can sometimes affect the neighboring genes too;
- Not very suitable for highly polyploid cell lines.
Nevertheless, these limitations can be minimized with bioinformatic, genetic and bibliographic analyses. Moreover, an alternative is represented by functional knockouts, in which the use of CRISPR/Cas9 or other genome editing methods allows the introduction of a point mutation to produce an inactive protein.
The generation of KO cell lines requires several steps, summarized by the following image.

Clone screening is needed to select the clones that correctly integrated the plasmid, which come in the end in a small number, reducing the efficiency of the process. An alternative is to use the newest technologies in genome editing, such as TALEN and ZNF (besides the most diffused CRISPR/Cas9, which is discussed in the Genome Editing section of this Wiki: http://cmb.i-learn.unito.it/mod/wiki/view.php?pageid=114).